
Florian Gerber
Data Scientist at Inter IKEA Group
florian.gerber@inter.ikea.com
I apply and develop cutting-edge machine learning methods for the analysis of large datasets.
Curriculum vitae
Experience |
|
|---|---|
| 2021 |
Data scientist at Inter IKEA Group, Supply Chain Design & Planning, Pratteln, Switzerland |
| 2018 |
PostDoc at the Applied Mathematics and Statistics Department, Colorado School of Mines, Golden CO, USA |
| Visiting scientist at the National Center for Atmospheric Research, Boulder CO, USA | |
| 2013— |
PhD assistant and PostDoc Department of Mathematics, University of Zurich, Switzerland |
| 2012— |
Junior statistician at the Institute of Social and Preventive Medicine, University of Bern, Switzerland |
| 2011— |
External statistics and R programming consultant for Novartis Pharma AG, Basel, Switzerland |
Education |
|
|---|---|
| 2017: | PhD in Applied Statistics, University of Zurich, Switzerland |
| 2013: | MSc in Biostatistics, University of Zurich, Switzerland |
| 2010: | BSc in Mathematics with minor in Philosophy, University of Bern, Switzerland |
| 2009: | Certificate in Interdisciplinary Studies in Ecology, University of Bern, Switzerland |
| 2006: | Matura, Gymnasiums Biel-Seeland, Switzerland |
Research
- develop cutting-edge machine learning methods for large datasets
- high-performance and cloud computing, statistical software
- Gaussian processes, Bayesian inference, neural networks, deep learning
- image processing for remote sensing, climate model, and medical applications
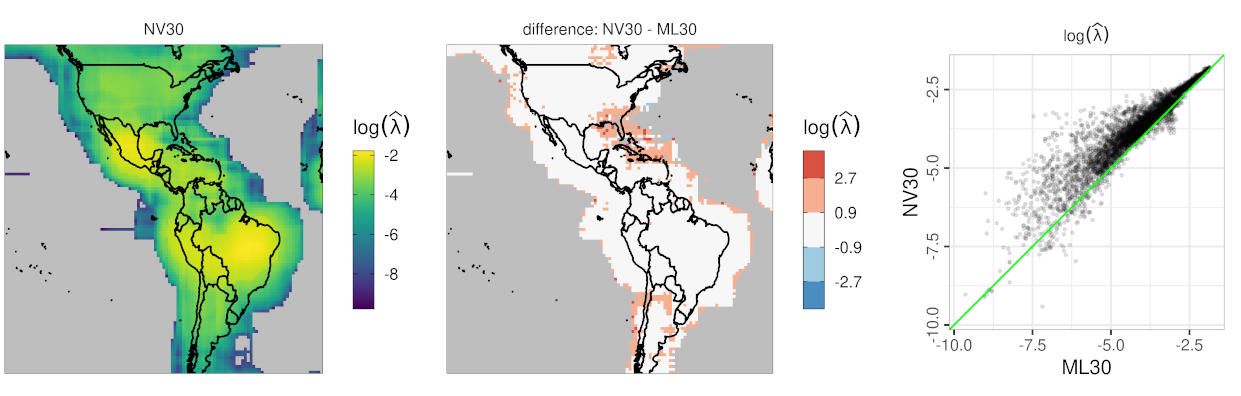
I. Use neural networks for fast covariance parameter estimation
- reduces the computing time for covariance parameter estimation of Gaussian process models by at least a factor of 100
- the TensorFlow implementation features a customized data generator defined using the Python API
- data illustration using climate model output from the LENS project
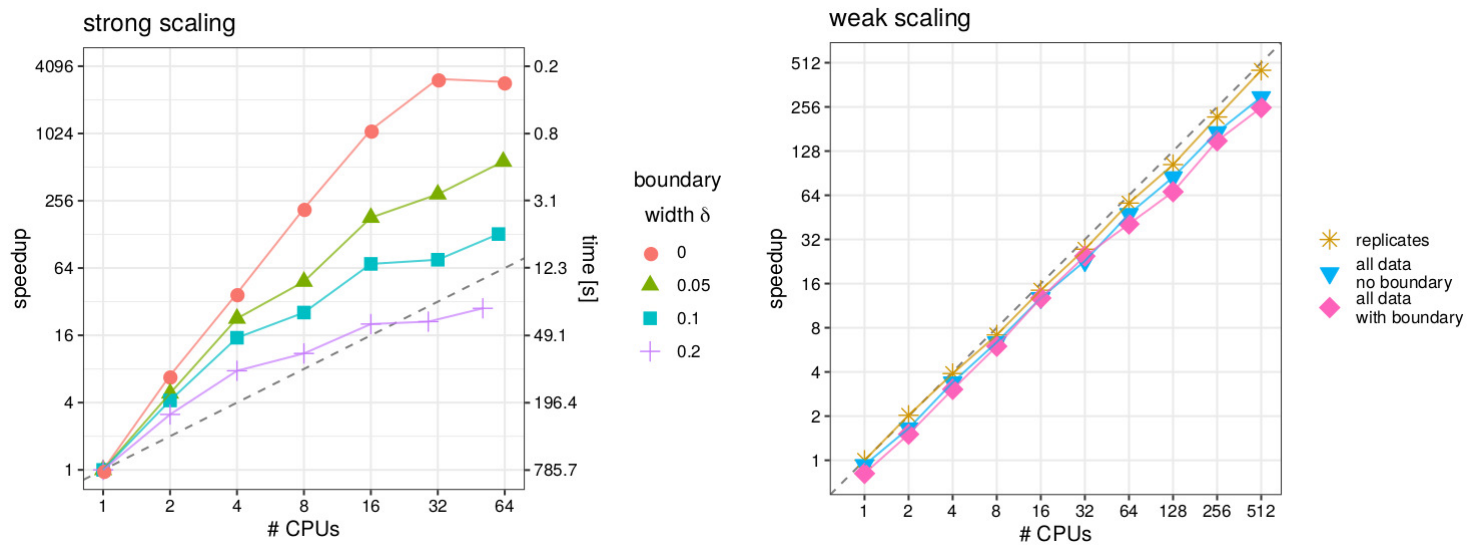
II. Parallel computing for Gaussian process models
- a Gaussian process model is fitted to a spatial dataset with 5 million observations
- high-performance computing with 512 CPUs in parallel is used
- uses cross-validation and the division of the domain in overlapping subsets
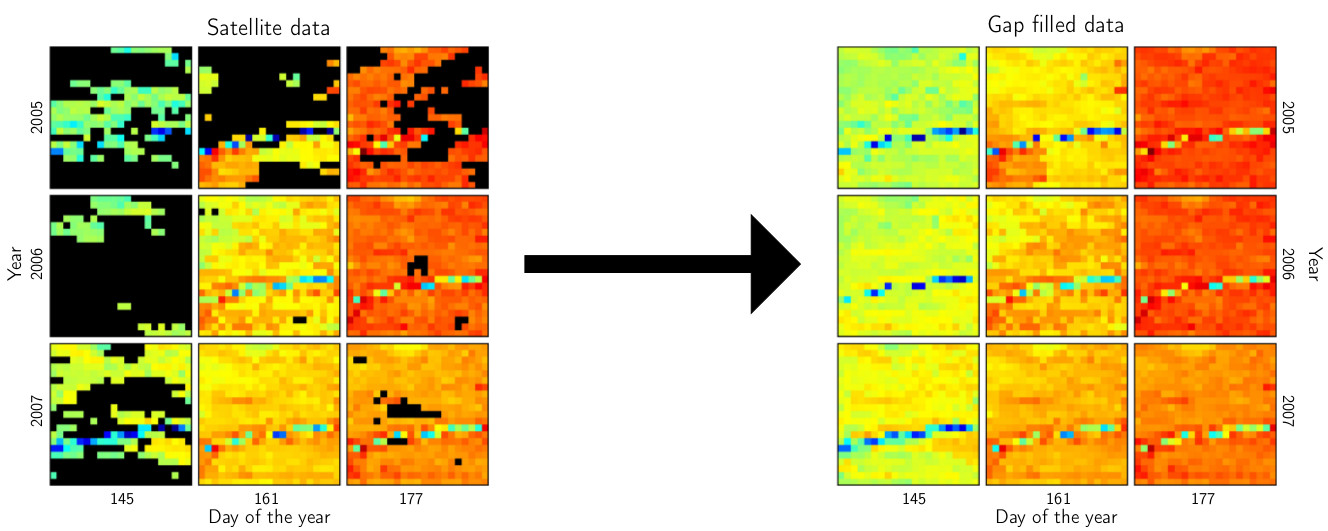
III. Predicting missing values in spatio-temporal remote sensing data
- predictions are obtained by selecting subsets, sorting procedures, and quantile regression
- the workload can be distributed to several computers
- the method is available in the R package gapfill available on CRAN
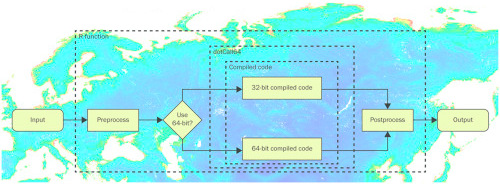
IV. 64-bit sparse matrices with the R package spam
- the R packages spam and spam64 provide methods for sparse matrices with more than 2^31 non-zero elements
- the R package dotCall64 provides an enhanced interface to link R with compiled code
- the new functionality of the R packages is illustrated with a non-stationary spatial model
Gerber F., Mösinger K, Furrer R. dotCall64: An Efficient Interface to Compiled C/C++ and Fortran Code Supporting Long Vectors, SoftwareX, 2018
V. Bayesian inference and medical applications
- assessment of inference methods for Bayesian hierarchical models
- spatial models for areal count data
- R package gsbDesign for operating characteristics of Bayesian clinical trial designs
Gerber F., Gsponer T. gsbDesign: An R Package for Evaluating the Operating Characteristics of a Group Sequential Bayesian Design, Journal of Statistical Software, 2016
Gsponer T., Gerber F., Bornkamp B., Ohlssen D., Vandemeulebroecke M., Schmidli H. Bayesian Group Sequential Designs for Clinical Trials and their Operating Characteristics, Pharmaceutical Statistics, 2014
Software
R packages on CRAN |
|---|
| optimParallel:
Parallel versions of the L-BFGS-B optimizer
|
| gapfill:
Fill missing values in satellite data
|
| gsbDesign:
Operating characteristics for clinical, Bayesian trials designs
|
| spam:
Sparse matrix algebra
|
| spam64:
64-bit extension of spam
|
| dotCall64:
Enhanced foreign function interface supporting long vectors
|
| fields:
Tools for spatial data
|
| vcd:
Tools for categorical data
|
Python package on PyPI |
optimparallel:
Parallel versions of L-BFGS-B optimization method in scipy
 |
Shiny R web application |
| Shut the Box: A variant of the dice game |
R packages on CRAN |
||
|---|---|---|
| optimParallel | Parallel versions of the L-BFGS-B optimizer | |
| gapfill | Fill missing values in satellite data | |
| gsbDesign | Operating characteristics for clinical, Bayesian trials designs | |
| spam | Sparse matrix algebra | |
| spam64 | 64-bit extension of spam | |
| dotCall64 | Enhanced foreign function interface supporting long vectors | |
| fields | Tools for spatial data | |
| vcd | Tools for categorical data | |
Python package on PyPI |
||
| optimparallel | Parallel versions of L-BFGS-B optimization method in scipy |  |
Shiny R web application |
||
| Shut the Box | A variant of the dice game | |
Publications
[16] Gerber, F. and Nychka, D. W. Fast covariance parameter estimation of spatial Gaussian process models using neural networks. Stat, 2022. doi: 10.1002/sta4.382
[15] Roman, F. Gerber, F., Schmid, B, and Furrer, R. Identification of dominant features in spatial data. Spatial Statistics, 2021. doi: 10.1016/j.spasta.2020.100483
[14] Gerber, F., and Nychka, D. W. Parallel cross-validation: a scalable fitting method for Gaussian process models. Computational Statistics & Data Analysis, 2021. doi: 10.1016/j.csda.2020.107113
[13] Gerber, F., and Furrer, R. optimParallel: An R package providing a parallel version of the L-BFGS-B optimization method. The R Journal, 2019. doi: 10.32614/RJ-2019-030
[12] Heaton, M. J., Datta, A., Finley, A., Furrer, R., Guinness, J., Guhaniyogi, R., Gerber, F., Gramacy, R. B., Hammerling, D., Katzfuss, M., Lindgren, F., Nychka, D. W., Sun, F., and Zammit-Mangion, A. A case study competition among methods for analyzing large spatial data. Journal of Agricultural, Biological and Environmental Statistics, 2018. doi: 10.1007/s13253-018-00348-w
[11] Warren, B. H., Hagen, O., Gerber, F., Thébaud, C., Paradis, E., and Conti, E. Evaluating alternative explanations for an association of extinction risk and evolutionary uniqueness in multiple insular lineages. Evolution, 2018. doi: 10.1111/evo.13582
[10] Gerber, F., Mösinger, K., and Furrer, R. dotCall64: An efficient interface to compiled C/C++ and Fortran code supporting long vectors. SoftwareX, 2018. doi: 10.1016/j.softx.2018.06.002
[9] Gerber, F., de Jong, R., Schaepman, M. E., Schaepman-Strub, G., and Furrer, R. Predicting missing values in spatio-temporal remote sensing data. IEEE Transactions on Geoscience and Remote Sensing, 2018. doi: 10.1109/TGRS.2017.2785240
[8] Gerber, F., Mösinger, K., and Furrer, R. Extending R packages to support 64-bit compiled code: An illustration with spam64 and GIMMS NDVI3g data. Computers & Geosciences, 2017. doi: 10.1016/j.cageo.2016.11.015
[7] Gerber, F. and Gsponer, T. gsbDesign: An R package for evaluating the operating characteristics of a group sequential Bayesian design. Journal of Statistical Software, 2016. doi: 10.18637/jss.v069.i11
[6] Bratulic, S., Gerber, F., and Wagner, A. Mistranslation drives the evolution of robustness in tem-1 β-lactamase. Proceedings of the National Academy of Sciences, 2015. doi: 10.1073/pnas.1510071112
[5] Gerber, F. and Furrer, R. Pitfalls in the implementation of Bayesian hierarchical modeling of areal count data: An illustration using BYM and Leroux models. Journal of Statistical Software, 2015. doi: 10.18637/jss.v063.c01
[4] Bashir, T., Sailer, C., Gerber, F., Loganathan, N., Bhoopalan, H., Eichenberger, C., Grossniklaus, U., and Ramamurthy, B. Hybridization alters spontaneous mutation rates in a parent-of-origin-dependent fashion in Arabidopsis. Plant Physiology, 165(1):424–437, 2014. doi: 10.1104/pp.114.238451
[3] Gsponer, T., Gerber, F., Bornkamp, B., Ohlssen, D., Vandemeulebroecke, M., and Schmidli, H. A practical guide to Bayesian group sequential designs. Pharmaceutical Statistics, 2014. doi: 10.1002/pst.1593
[2] Gerber, F., Marty, F., Eijkel, G. B., Basler, K., Brunner, E., Furrer, R., and Heeren, R. M. A. Multiorder correction algorithms to remove image distortions from mass spectrometry imaging data sets. Analytical Chemistry, 2013. doi: 10.1021/ac402018e
[1] Wandeler, G., Gerber, F., Rohr, J., Chi, B., Orrell, C., Chimbetete, C., Prozesky, H., Boulle, A., Hoffmann, C., Gsponer, T., Fox, M., Zwahlen, M., Egger, M., and IeDEA S. A. Tenofovir or Zidovudine in second-line antiretroviral therapy after Stavudine failure in Southern Africa. Antiviral Therapy, 2013. doi: 10.3851/imp2710
